| 图片: | |
|---|---|
| 名称: | |
| 描述: | |
- 请欣赏教学片
-
Polyomavirus Inclusion Bearing “Decoy” Cells
Beginning in 1945, George Papanicolaou stressed the usefulness of urine cytology examination and the “Papanicolaou stain” for the diagnostic evaluation of cellular elements in voided urine specimens. This technique rapidly gained worldwide acceptance since it provided an easy, reliable, and inexpensive clinical tool. Approximately forty years ago, Koss and colleagues described polyomavirus inclusion bearing cells for the first time in urine cytology specimens.15 They coined the term “decoy cells” to alert pathologists not to misdiagnose viral inclusion bearing cells as malignant cancer cells.
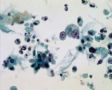
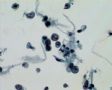
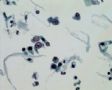
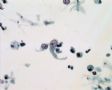
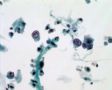
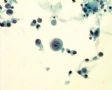
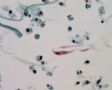
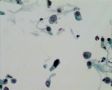
Decoy-Cells, Morphology and CharacterizationThe name “decoy cell” is a descriptive term for epithelial cells with intranuclear viral inclusion bodies that can have different phenotypes (types 1-4) depending upon the state of viral replication and maturation as well as the state of cellular preservation. The order in which the various phenotypes may occur during intranuclear viral assembly is unclear. Hybrid forms representing transitions between the different phenotypes are frequently found in the same specimen. Most common are classic decoy cells characterized by large, homogenous, amorphous ground-glass like intranuclear inclusion bodies and a condensed rim of chromatin (type 1) (fig. 1). Sometimes, decoy cells reveal granular intranuclear inclusions surrounded by a clear halo, i.e., cytomegalovirus (CMV)-like (type 2) (fig. 2). Occasionally, multinucleated decoy cells with granular chromatin are detected (type 3) (fig. 3). Type 4 decoy cells show vesicular nuclei, often with clumped chromatin and nucleoli (fig. 4). Koss called these latter inclusions the “empty post-inclusion stage”.15 Types 3 and 4 decoy cells are especially prone to misinterpretation as “cancer cells”. Although the nuclear features are most characteristic, many decoy cells additionally show a typical eccentric cytoplasm resembling the tail of a comet (termed “comet cells” by some, fig. 1 and fig.2). onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913297);">16
Decoy cells mostly contain polyomaviruses of the BK strain or less commonly of the JC strain. Rarely, also adenoviruses may be found (Table 1). onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913370);">5, onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913384);">17, onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913399);">18 Immunohistochemical and electron microscopical analyses can easily be used to verify the presence of viruses and to identify the virus families. In general, most types 1 through 4 intranuclear inclusion bodies in decoy cells give a positive staining reaction with a commercially available antibody detecting the simian virus “SV-40 T antigen” which is common to all known polyomavirus strains pathogenic in humans (i.e., BK-, JC-, SV-40 strains (fig. 5); see appendix for staining protocols). onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913425);">7,19 Of note: since the large T antigen is only expressed in abundancy during the early phases of viral replication, decoy cells with late stages of polyomavirus assembly may be “T antigen” negative. Using BK-virus specific antibodies or PCR techniques, most decoy cells contain polyoma-BK-virus particles. onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913267);">5 Immunohistochemistry can also help to identify adenovirus containing decoy cells. Electron microscopy is well suited to detect polyomaviruses and adenoviruses based on their characteristic size of 40—50 nm and 80 nm, respectively (fig. 6). Ultrastructural analysis, however, is not suited to distinguish between different polyoma- or adenovirus strains. Although productive infections with cytomegalovirus, herpes simplex virus or human papillomavirus can show nuclear abnormalities including viral inclusion bodies, typical “decoy cells” as described above are generally not found in the urine in these infections (Table 1).
Decoy-Cells, OriginThe origin of decoy cells cannot be easily discerned based on morphologic grounds. It seems likely to us that they would commonly originate from the urothelium, in particular, in healthy and asymptomatic patients (fig. 7a). onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913598);">5, onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913612);">7 This assumption is based on the observation that the urothelium often harbors latent BK-virus infections (approximately 50% of individuals, personal observation). The replication of polyomaviruses is most pronounced in the superficial transitional cell layer, i.e., in umbrella cells, which can easily be shed. onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913627);">9 As mentioned above, decoy cell shedding is often asymptomatic and renal function remains unaltered. Polyomavirus inclusion bearing cells are never seen in native kidneys of immune competent patients further arguing for an extra (renal) parenchymal origin of decoy cells found in the urine of healthy individuals.
In contrast, in immunocompromised patients BKN is characterized by intra-renal replication of BK viruses and kidney dysfunction. The morphological signs of viral replication in renal tubular epithelial cells in cases of BKN are very similar to those seen in transitional cells and decoy cells (see Chapter 12: “Latent and Productive Polyomavirus Infections of Renal Allografts: Morphological, Clinical and Pathophysiological Aspects”). Thus, in cases of BKN, decoy cells likely also originate from the renal parenchyma. onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913478);">7, onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913666);">9,19 It is tempting to speculate that BKN may be caused by an ascending route of infection with spreading of polyomavirus replication from transitional cells to collecting ducts and proximal tubular epithelial cells in some patients in whom risk factors provide the right window of opportunity (fig. 7b and fig. 7c). onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913711);">7 However, this hypopthesis has not yet been proven.
Decoy-Cells Versus Malignant Tumor CellsOne of the most important challenges, already stressed by Koss and his colleagues, is to properly identify decoy cells and to avoid their misinterpretation as “malignant tumor cells”. 15Sound knowledge of the various phenotypes of viral inclusion bodies and the utilization of immunohistochemical and electron microscopic analyses should generally lead to their proper identification. In cases of polyomavirus activation and replication, the evaluation of “atypical cells” with proliferation markers or by DNA image cytometry can be misleading. Polyomaviruses require the “machinery” of the host cells for viral amplification. Thus, immunohistochemical stains to detect “proliferation associated antigens”, such as antibodies directed against proliferating cell nuclear antigen (PCNA), KI-67 or MIB-1, give strong signals in decoy cells and inclusion bearing transitional cells. Such staining profiles should not be misinterpreted as a sign of marked “cell ” proliferation, but rather indicate the replication of viral DNA. Accordingly, DNA cytometry/histograms of decoy cells invariably show aneuploidy due to the viral DNA content (fig. 8). In contrast, FISH with chromosome enumeration probes and single locus specific identifiers (9p21) (UroVision,TM Vysis Inc., Downers Grove IL), can reliably demonstrate normal chromosome and gene copy numbers (fig.8). 20 The FISH profile clearly identifies decoy cells as “benign” and distinguishes them from cancer cells. onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913798);">21
Decoy-Cells, DetectionDecoy cells can best be identified and quantified in standard alcohol fixed and Papanicolaou stained urine cytology specimens from either smeared or cytocentrifuged (i.e., cytospin) urine samples. In addition, the recently introduced monolayer technique is also feasible since it provides excellent preservation of nuclear details (Cytyc Inc., Boxborough, MA, and Tripath Imaging Inc., Burlington, NC). The second morning midstream voided urine specimen is best suited as the high level of cellular degeneration severely limits the first morning specimen. A few important considerations in the handling of urine specimens are the following: (1) Fresh urine specimens should be promptly transported to the cytology laboratory for immediate processing. (2) An alternative and more frequently used method is the fixation of the urine with an equal volume of 50-70% ethyl alcohol, preferably with added 2% Carbowax (polyethylene glycol). This procedure can be performed beside or in the laboratory when delayed specimen handling is anticipated. (3) Conventional cytospin or smear preparations should be obtained immediately after the specimen is received in the cytology laboratory. (4) The use of coated glass slides is recommended for adequate adherence of the cellular and noncellular elements to the slide surface. 22 It should be noted that specific guidelines for adequacy of voided urine specimens at the time of cytologic interpretation have not been firmly established and much is still dependent on the experience level of the pathologist. 22 Decoy cells can also be detected in the unstained urine sediment using phase contrast microscopy. onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913970);">8, onmouseout="PopUpMenu2_Hide(); return false;" href="javascript:PopUpMenu2_Set(Menu_id913984);">23 This technique, however, requires great experience and the quantification of decoy cells is tricky. We, therefore, recommend the analysis of standard Papanicolaou stained cytology specimens as described above.

- “人生没有彩排,每一天都是现场直播”