| 图片: | |
|---|---|
| 名称: | |
| 描述: | |
- B3137Dr.cqzhao推荐的一篇好文章:Basal-like and triple-negative breast cancers
Review
Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists
Sunil Badve1, David J Dabbs2, Stuart J Schnitt3, Frederick L Baehner4, Thomas Decker5, Vincenzo Eusebi6, Stephen B Fox7, Shu Ichihara8, Jocelyne Jacquemier9, Sunil R Lakhani10, Jose´ Palacios11, Emad A Rakha12, Andrea L Richardson13, Fernando C Schmitt14, Puay-Hoon Tan15, Gary M Tse16, Britta Weigelt17, Ian O Ellis12 and Jorge S Reis-Filho18
1Clarian Pathology Lab, Department of Pathology and Internal Medicine, Indiana University, Indianapolis, IN, USA; 2Department of Pathology, University of Pittsburgh Medical Center, Pittsburgh, PA, USA; 3Department of Pathology, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA, USA; 4Department of Anatomic Pathology, University of California, San Francisco, CA, USA; 5Breast-Screening-Pathology, Gerhard Domagk-Institute of Pathology, Reference Centre Mu¨nster, University Hospital Mu¨nster, Mu¨nster, Germany; 6Sezione Anatomia Istologia e Citologia Patologica ‘M Malpighi’, Universita`-ASL Ospedale Bellaria, Bologna, Italy; 7Pathology Department, Peter MacCallum Cancer Centre, East Melbourne, Victoria, Australia; 8Department of Pathology, Nagoya Medical Center, Nagoya, Japan; 9Unite´ d’Anatomie et de Cytologie Pathologiques, Institut Paoli-Calmettes, Marseille, France; 10Molecular and Cellular Pathology, University of Queensland Centre for Clinical Research, School of Medicine and Pathology Queensland and The Royal Brisbane andWomen’s Hospital, Brisbane, Australia; 11Servicio de Anatomı´a Patolo´gica, Hospital Universitario Virgen del Rocı´o, Sevilla, Spain; 12Department of Histopathology, Nottingham City Hospital NHS Trust, Nottingham University, Nottingham, UK; 13Department of Pathology, Brigham and Women’s Hospital, Boston, MA, USA; 14Medical Faculty, Institute of Molecular Pathology and Immunology (IPATIMUP), University of Porto, Porto, Portugal; 15Department of Pathology, Singapore General Hospital, Singapore, Singapore; 16Department of Anatomical and Cellular Pathology, Prince of Wales Hospital, The Chinese University of Hong Kong, Hong Kong, China; 17Signal Transduction Laboratory, Cancer Research UK London Research Institute, London, UK and 18The Breakthrough Breast Cancer Research Centre, Institute of Cancer Research, London, UK Breast cancer is a heterogeneous disease encompassing a variety of entities with distinct morphological features and clinical behaviors. Although morphology is often associated with the pattern of molecular aberrations in breast cancers, it is also clear that tumors of the same histological type show remarkably different clinical behavior. This is particularly true for ‘basal-like cancer’, which is an entity defined using gene expression analysis. The purpose of this article was to review the current state of knowledge of basal-like breast cancers, to discuss the relationship between basal-like and triple-negative breast cancers, and to clarify practical implications of these diagnoses for pathologists and oncologists.
Modern Pathology (2011) 24, 157–167; doi:10.1038/modpathol.2010.200; published online 12 November 2010
Keywords: basal-like; breast cancer; definition; practical review; triple-negative
Breast cancer is a heterogeneous disease, and this term encompasses a variety of entities with distinct morphological features and clinical behaviors. In recent years, it has become apparent that this diversity is the result of distinct genetic, epigenetic, and transcriptomic alterations.1–5 Although morphology is often associated with the pattern of molecular aberrations in breast cancers,5 it is also clear that tumors of the same histological type show remarkably different clinical behavior. This is most evident in invasive ductal carcinomas of no special type (IDC-NST), where even tumors of the same histological grade may have distinct outcomes and dramatically different responses to systemic therapy.2,3 Using high-throughput technologies, particularly microarray analysis, several groups have proposed a new taxonomy for breast cancer based on their molecular features. The gene expression microarraybased class discovery studies pioneered by the Stanford group have led to the identification of at least five molecular breast cancer subtypes: luminal A, luminal B, normal breast-like, HER2, and basallike. 6–11 Although based on the analysis of a limited number of samples and with somewhat different definitions for the various molecular groups in these studies, this approach to the classification of breast cancer has captured the attention of oncologists, pathologists, and scientists alike.
It should be noted, however, that this taxonomy has identified subgroups of breast cancer that were to some extent already known, and that the stability of the assignments of molecular subtypes by microarray-based methods has been called into question.12–14 Indeed, the most robust distinction observed by microarray analysis is between the transcriptome of estrogen receptor-positive (ERt) and ER-negative (ER_) breast cancers.
Among the molecular subtypes of breast cancer identified through gene expression profiling studies, none has generated as much interest or controversy as the basal-like group. The purpose of this article was to review the current state of knowledge of basal-like breast cancers, to discuss the relationship between basal-like and triple-negative breast cancers, and to clarify practical implications of these diagnoses for pathologists and oncologists.
What is a basal-like breast cancer?
The characteristics of basal-like breast cancer have been extensively reviewed in the past 18 months.10,14–16 It should be noted that there is still no internationally accepted definition for basal-like breast cancers and how best to define these tumors is a matter of controversy and ongoing debate. Some groups have used microarray-based expression profiling to define basal-like breast cancers, whereas others have used panels of immunohistochemical markers as surrogates. However, direct comparisons between the proposed immunohistochemical markers and the microarray-defined molecular subtypes are scarce.17,18 Immunohistochemical marker panels that have been proposed to define basal-like breast cancers include: (1) lack of ER, PR, and HER2 expression (‘triple-negative’ immunophenotype); (2) expression of one or more high-molecular-weight/ basal cytokeratins (CK5/6, CK14, and CK17); (3) lack of expression of ER and HER2 in conjunction with expression of CK5/6 and/or epidermal growth factor receptor (EGFR);17 and (4) lack of expression of ER, PR, and HER2 in conjunction with expression of CK5/6 and/or EGFR.19
Despite the different definitions for basal-like breast cancers, it has been demonstrated that these tumors have distinctive clinical presentations,20 histological features,18,21 response to chemotherapy, 22–28 sites of distant relapse, and outcome.7,9,29–31 In brief, basal-like tumors comprise a heterogeneous group that accounts for up to 15% of all breast cancers, affect younger patients, are more prevalent in African-American women, and often present as interval cancers. Histologically, the majority of basal-like breast cancers is of IDC-NST type, high histological grade, and characterized by exceptionally high mitotic indices, the presence of central necrotic or fibrotic zones, pushing borders, conspicuous lymphocytic infiltrate, and typical/atypical medullary features (Figure 1).18,21,32 The similarity of these features with those of human papilloma virus-induced squamous cell carcinoma of the head and neck has led to the identification of the role of retinoblastoma gene (RB1) in these tumors.33 However, not all basal-like cancers are of the IDC-NST type; the majority of medullary and atypical medullary, 4,34–36 metaplastic,4,37,38 secretory,39 myoepithelial, and adenoid cystic carcinomas4,40 of the breast also show a basal-like phenotype.5 More recently, a subgroup of lobular carcinomas has been shown to express high-molecular-weight cytokeratins,41 however it remains to be determined whether these cases truly show a basal-like transcriptome. The majority of basal-like breast cancers lack or show low levels of ER and PR, lack HER2 protein overexpression and HER2 gene amplification, whereas they express genes and proteins usually found in ‘basal’/myoepithelial cells of the normal breast including high-molecular-weight cytokeratins (5/6, 14 and 17),17,19,21,42 P-cadherin,43 caveolins 1 and 2,44,45 nestin,46 aB crystallin,47,48 CD109,49,50 and EGFR17 and, in a minority of cases, harbor EGFR gene amplification51 or aneusomy.52 p53 immunohistochemical expression or TP53 gene mutations is observed in up to 85% of cases,53,54 and alterations of the pRB and p16 G1/S cell-cycle checkpoint are remarkably prevalent in these cancers.33,55 A recent study demonstrated that approximately 30% of basal-like breast cancers concurrently show lack of pRB expression, overexpression of p16 and p53 immunoreactivity (pRB_/p16t/p53t), whereas this profile was rarely seen in tumors of other molecular subtypes.33 Basal-like cancers show remarkably high proliferation indices as defined by mitotic counting or by Ki67 labeling index.26,33,56
Of interest, basal-like breast cancers, unlike ‘basal’/myoepithelial cells of normal breast, almost uniformly express cytokeratins 8 and/or 18, calling into question the initial histogenetic implications of the microarray-based taxonomy of breast cancers that suggested that basal-like cancers would arise from basal/myoepithelial cells.5,14 This has been emphasized in a recent study, which suggested that at least a subgroup of basal-like breast cancers may originate from luminal progenitors rather than basal/ myoepithelial cells of the breast,57 and confirmed by the results of conditional mouse models.58 In this context, it is important to note that histogenesis and differentiation are two distinct processes although often mistakenly used as synonyms.
What is a triple-negative breast cancer?
In contrast to the controversy regarding the definition of basal-like breast cancers, there is uniform agreement that triple-negative cancers are defined as tumors that lack ER, PR, and HER2 expression. These tumors account for 10–17% of all breast carcinomas,1,24,28,59–65 depending on the thresholds used to define ER and PR positivity and the methods used for HER2 assessment. Future studies are likely to produce slightly different prevalence rates for triple-negative breast cancers given the change in the definition of HER2 and hormone receptor positivity according to the ASCO/CAP guidelines. 66,67 Despite these definitional issues, the clinical interest in these tumors stems from the lack of tailored therapies for this group of breast cancer patients and the overlap with the profiles of basallike cancers.
The main characteristics of triple-negative cancers that have emerged from the literature illustrate their similarities to basal-like cancers, including the fact that they more frequently affect younger patients (o50 years),24,30,31,59,61,62 are more prevalent in African-American women,62–64 often present as interval cancers, and are significantly more aggressive than tumors of other molecular subtypes. 24,28,59,61–63 This aggressiveness is best exemplified by the fact that the peak risk of recurrence is between the first and third years and the majority of deaths occur in the first 5 years following therapy.59,61 Patients with triple-negative cancers,59,63 similar to those with basal-like cancers, 42 have a significantly shorter survival following the first metastatic event when compared with those with non-basal-like/non-triple-negative controls.
Are basal-like and triple-negative cancers synonymous?
As should be evident from the foregoing discussion, although there are numerous similarities between basal-like and triple-negative breast cancers, these two terms are not synonymous, despite the fact that some have previously used these terms interchangeably. 10,15,53,68,69 It is true that the majority of triplenegative cancers are of basal-like phenotype22,60,68 and the majority of tumors expressing ‘basal’ markers are triple-negative.16,17,19,53,60,70 However, not all basal-like cancers determined by gene expression profiling lack ER, PR and HER2,10,12,15,16,68,70–74 and conversely not all triplenegative cancers show a basal-like phenotype by expression array analysis. For example, Bertucci et al70 showed that only 71% of triple-negative cancers were of basal-like subtype by gene expression profiling and that only 77% of molecular basallike tumors were triple-negative. Similar results were observed by de Ronde et al74 and Parker et al (Supplementary Table 1 of Parker et al11), where 8–29% of triple-negative cancers did not show a basal-like subtype by expression array analysis and 18–40% of basal-like breast cancers defined by gene expression profiling did not harbor a triple-negative phenotype. Further, there is a significant number of triple-negative cancers that do not express basal markers and are classified as normal breast-like (probably an artifact of gene expression profiling due to samples with disproportionately high content of stromal and normal breast epithelial cells),11,75,76 molecular apocrine77,78 (tumors with androgen receptor pathway activation, although a substantial proportion of these tumors may be classified as of HER2 subtype78 using other molecular classification systems) or claudin-low79,80 (cancers with transcriptomic features suggestive of epithelial to mesenchymal transition and reported to be enriched for the so-called ‘cancer stem cells’) subtype by gene expression profiling (for review, see Weigelt et al12). Apart from the more heterogeneous transcriptome, triple-negative cancers also show more varied histological features. Indeed, up to 10% of triplenegative tumors were reported to be of grade I in one study.59 However, numerous other studies have failed to identify any grade I breast cancers with a triple-negative phenotype. Furthermore, other histological special types of breast cancer that do not show a basal-like phenotype by transcriptomic analysis have been shown to occasionally express a triple-negative phenotype, including apocrine carcinomas, pleomorphic lobular carcinomas, and some mixed duct-lobular cancers.1,15,69 Taken together, these results are in accord with the concept that the triple-negative phenotype is not an ideal surrogate marker for basal-like breast cancers.60,70,81
Relationship between basal-like breast cancer and BRCA1 germ-line mutations
There is increasing evidence to suggest a link between BRCA1 pathway and basal-like breast cancers.82,83 The majority of tumors arising in BRCA1 germ-line mutation carriers, in particular those diagnosed before 50 years of age, have morphological features similar to those described in basal-like cancers84,85 and show a basal-like phenotype as defined by immunohistochemistry86,87 or expression arrays.8
Both basal-like breast cancers and tumors arising in BRCA1 germ-line mutation carriers show a peculiar pattern of cell-cycle protein expression; 54,84,88,89 both rarely harbor CCND1 gene amplification; 54,88 however, they express significantly lower levels of p27,84,89 and higher levels of Skp2,84,89 cyclin E,84,89 and caspase-3,89 when compared with sporadic breast carcinomas and BRCA2 mutation tumors.
Although they lack BRCA1 somatic mutations, sporadic basal-like cancers show similar molecular genetic profiles to tumors arising in BRCA1 mutation carriers.90–94 This may be in part due to the presence of a dysfunctional BRCA1 pathway in these tumors.32,82,83 BRCA1 gene promoter is methylated in 460% of medullary95,96 and metaplastic32 breast cancers of basal-like phenotype. Sporadic invasive ductal carcinomas with basal-like phenotype express ID4, a negative regulator of BRCA1,97,98 at significantly higher levels than grade-matched controls.32 This mechanism may account for the low levels of BRCA1 expression in sporadic basal-like carcinomas of ductal morphology. Importantly, recent studies have demonstrated that BRCA1 gene silencing leads to downregulation of ER99 and upregulation of genes considered to be markers of basal-like cancers,100 including CK5, CK17, and P-cadherin. In contrast, reconstitution of BRCA1 in ER_ BRCA1 mutant cell lines has been shown to lead to upregulation of ER and downregulation of CK5, CK17, and P-cadherin.99,100 Taken together, BRCA1 dysfunction appears to be one of the drivers of basal-like breast cancers and of a subgroup of triple-negative tumors.
Precursors of basal-like and triple-negative cancers
A group of high-grade DCIS lacking ER, PR and HER2, and expressing ‘basal’ markers has been identified.101–107 However, it should be noted that its prevalence is lower than that of invasive triplenegative and basal-like breast cancers and that triple-negative and basal-like cancers often lack an overt in situ component. Whether this is the result of basal-like and triple-negative breast cancers progressing rapidly from DCIS to invasive cancer and/or obliterating the DCIS precursor from which they arose remains a matter of speculation.
The majority of invasive cancers developing in microglandular adenosis is of triple-negative phenotype108– 112 and show metaplastic elements or is of adenoid cystic morphology.109–112 It has been recently shown that microglandular adenosis may be a nonobligate precursor of triple-negative and basallike breast cancers.108,113 A stepwise progression in the number of gross chromosomal changes from microglandular adenosis to invasive carcinoma has been observed.108 However, given the rarity of microglandular adenosis, it is unlikely to be the precursor lesion for most triple-negative cancers.
Clinical behavior of basal-like and triple-negative breast cancers
Basal-like and triple-negative breast cancers, as defined by microarrays or by immunohistochemical surrogates, have been shown to have a more aggressive clinical behavior.17,114,115 In fact, some studies have demonstrated that expression of basal keratins is a prognostic factor independent of tumor size, grade, and lymph node status.114 However, when compared with either ER_ non-basal-like cancers72 or grade-matched non-basal-like cancers,42 carcinomas with a basal-like phenotype are not associated with a poorer outcome in some studies, whereas a more adverse prognosis is observed in others.19,116 The pattern of metastatic spread of tumors with a basal-like phenotype seems to be different from that of non-basal-like cancers: they are reported to less frequently disseminate to axillary nodes and bones42,117 and to favor a hematogenous spread,42,117–119 with a peculiar proclivity to develop metastatic deposits in the brain and lungs.120 It should be noted that patients with triple-negative and basal-like cancers tend to develop adverse events and die due to disease within the first 5–8 years after diagnosis. After the 8-year mark, the hazard rate for patients with grade 2 or ERpositive cancers is actually higher than that of patients with basal-like cancers. Finally, some tumors in the ‘basal’ group have a favorable prognosis, eg, adenoid cystic carcinomas40,121 and secretory carcinomas.39 This emphasizes that basal phenotype and bad behavior are not inextricably linked and serves to highlight the heterogeneous nature of basal-like carcinomas. It is controversial as to whether these tumors should be classified as the basal-like carcinomas or should represent a distinct group by themselves.
As to treatment response, 17–58% of patients with triple-negative breast cancers have been shown to have a pathological complete response after anthracycline- or anthracyclinettaxane-based neoadjuvant chemotherapy25,28,122,123 and 17% of triplenegative cancers have been shown to have a pathological complete response after neoadjuvant platinum-based chemotherapy.124 However, those who fail to achieve pathological complete response have a dismal outcome.25,28 It should be noted, however, that markers for the identification of patients with triple-negative and basal-like cancers that benefit most from chemotherapy remain to be defined. Recently, several groups have identified a subgroup of good prognosis ER_ cancers, encompassing a subgroup of triple-negative and basal-like tumors, which is characterized by the expression of an immune response module.125–128 This transcriptomic profile may prove helpful for the identification of patients with triple-negative and basal-like cancers that have a better outcome. In this context, the finding of higher expression of CT-X antigens (in particular MAGE and NY-ESO) in this subgroup opens up another potential avenue for therapy as vaccines against these antigens are already in clinical trial for lung cancer.129
Finally, as mentioned earlier, a group of basal-like and triple-negative breast carcinomas have been shown to have a dysfunctional BRCA1 pathway. 32,65,81–83 A significant number of these cases lack competent homologous recombination DNA repair. This subgroup may be amenable to specific therapeutic strategies such as inhibitors of the PARP enzyme.32,81–83,130,131 Consistent with this hypothesis, results of PARP inhibitor phase I and phase II clinical trials that have included patients with BRCA-deficient tumors have been encouraging132–134 and sustained responses in patients with BRCA1/2- deficient breast or ovarian metastatic cancers have been observed. Furthermore, preliminary results of a phase II clinical trial revealed that addition of BSI- 201, a PARP inhibitor, to gemcitabine/carboplatin chemotherapy led to a significantly increased clinical benefit and longer progression-free survival. 135 However, the specificity of this compound for PARP has yet to be fully established.
Given these exciting results, several clinical trials testing cross-linking agents (eg carboplatin and cisplatin) and PARP inhibitors in patients with BRCA1 germ-line mutations and sporadic basal-like breast cancers are currently ongoing (for a list of clinical trials, please see clinicaltrials.gov). If positive, these studies may render the identification of tumors lacking competent homologous recombination compulsory in our diagnostic practice.130
Practical implications of basal-like and triple-negative breast cancers
As outlined above, a diagnosis of triple-negative breast cancer has direct clinical implications. Given that these tumors lack expression of hormone receptors and lack HER2 protein overexpression and gene amplification, chemotherapy is the only modality of adjuvant systemic therapy currently available for patients with triple-negative disease. Furthermore, the current prognostic algorithms to define which patients should receive chemotherapy may not work optimally for patients with triplenegative breast cancer, given that the impact of size on the outcome of these patients appears to be significantly attenuated.136,137
At present, use of the term ‘basal-like breast cancer’ in diagnostic surgical pathology reports does not appear to be justified, as it does not lead to any direct clinical action. In fact, given the variations in definition of basal-like breast cancers, such a diagnosis may cause clinical confusion. For instance, according to some definitions, a breast cancer showing ER expression and expression of CK 5/6 or CK 14 would be classified as being of basallike phenotype. However, there are currently no data to indicate that patients with ERt breast cancers that show basal cytokeratin expression should be managed any differently than ERt, basal cytokeratin- negative breast cancers of equivalent size and stage (ie, with hormonal therapy, with or without chemotherapy). Likewise, patients with an HER2t breast cancer showing a basal-like phenotype would still receive trastuzumab and chemotherapy, similar to other patients with HER2t breast cancer. Thus, given that the term basal-like per se does not impact on clinical decision making, the inclusion of the term ‘basal-like breast cancer’ in pathology reports at present would be best avoided. However, it might be argued that subclassification of triple-negative tumors into subtypes using the Nielsen definition17 is of prognostic significance and is associated with specific patterns of metastatic dissemination. If one were to use such an approach, to ensure consistency it is recommended that tumors that express more than 1% CK5/6 or EGFR should be considered positive for these markers. There are also data to suggest that the CK5 antibody is superior to the CK5/6 antibody for identification of this type of basal cytokeratin.138 The role of CK14 in the subclassification of triple-negative cancers into basal-like and non-basal-like phenotypes is uncertain.
Perhaps more important than identifying the basal-like subgroup within triple-negative breast cancers is the identification of subgroups of triplenegative disease that are sensitive to specific systemic therapy regimens. Several groups are currently developing biomarkers for the identification of the subgroup of triple-negative cancers with dysfunctional homologous recombination DNA repair, given that these tumors are likely to show an exquisite sensitivity to PARP inhibitors. Furthermore, other promising targets for subgroups of basal-like cancers have recently emerged (eg, FGFR2, TRAIL, antiangiogenic agents). Therefore, the question that is germane for the management of breast cancer patients is the identification of predictive biomarkers to substratify patients with triple-negative cancers into groups that can be managed more efficaciously with specific systemic therapies.
It is also important to remember that the lack of overlap between categories identified by gene expression and immunohistochemistry methods is not restricted to basal-like and triple-negative tumors. Up to 25% of clinically ERt tumors are classified as of nonluminal subtype by gene expression methods.11 Similarly, nearly a third of the clinically HER2t (FISH and/or IHC) are not classified as belonging to HER2-enriched category.11 At least for now, treatment decisions are made on the basis of clinical (and not gene expression) assays.
Conclusions
Basal-like breast cancer is a heterogeneous group of tumors that is more prevalent in young and African- American patients and is generally associated with a poor outcome. Currently, although it is clearly important that triple-negative cancers be accurately identified in clinical practice for the purposes of management, there is no internationally accepted definition for basal-like cancers and still no clear clinical indication for the routine identification of these tumors as such.14
Given that basal-like breast cancers are heterogeneous regardless of the definition used, it is possible that in the next few years, markers that identify subgroups of basal-like or triple-negative cancers that respond to specific agents will become part of our diagnostic armamentarium. With the advent of massively parallel sequencing,139 which allows for the genome-wide quantitative and qualitative genomic and transcriptomic characterization of cancers, and the imminent death of microarrays,140,141 it is likely that the taxonomy of breast cancers will be revisited again.14,56 At that time, it is quite possible that more homogeneous molecular subgroups, their biological drivers, and therapeutic targets will be identified. Until then, it is essential that pathologists continue to strive toward providing optimal assessment of the histological features of breast cancers (including histological grade), as well as accurate determination of ER, PR, and HER2 status according to published guidelines66,67 as these factors remain the primary determinants of the use and type of systemic therapy for patients with invasive breast cancer.
References
1 Reis-Filho JS, Lakhani SR. Breast cancer special types: why bother? J Pathol 2008;216:394–398.
2 Geyer FC, Marchio C, Reis-Filho JS. The role of molecular analysis in breast cancer. Pathology 2009;41:77–88.
3 Correa-Geyer FC, Reis-Filho JS. Microarray-based gene expression profiling as a clinical tool for breast cancer management: are we there yet? Int J Surg Pathol 2009;17:285–302.
4 Weigelt B, Horlings HM, Kreike B, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol 2008;216:141–150.
5 Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol 2009;6:718–730.
6 Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000; 406:747–752.
7 Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001;98:10869–10874.
8 Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 2003;100:8418–8423.
9 Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 2006;7:96.
10 Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol 2008;26: 2568–2581.
11 Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–1167.
12 Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol 2010; 220:263–280.
13 Pusztai L, Mazouni C, Anderson K, et al. Molecular classification of breast cancer: limitations and potential. Oncologist 2006;11:868–877.
14 Gusterson B. Do ‘basal-like’ breast cancers really exist? Nat Rev Cancer 2009;9:128–134.
15 Rakha EA, Reis-Filho JS, Ellis IO. Impact of basal-like breast carcinoma determination for a more specific therapy. Pathobiology 2008;75:95–103.
16 Rakha EA, El-Sayed ME, Reis-Filho J, et al. Pathobiological aspects of basal-like breast cancer. Breast Cancer Res Treat 2008;113:411–422.
17 Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basallike subtype of invasive breast carcinoma. Clin Cancer Res 2004;10:5367–5374.
18 Livasy CA, Karaca G, Nanda R, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol 2006;19: 264–271.
19 Cheang MC, Voduc D, Bajdik C, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 2008;14:1368–1376.
20 Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006;295:2492–2502.
21 Fulford LG, Easton DF, Reis-Filho JS, et al. Specific morphological features predictive for the basal phenotype in grade 3 invasive ductal carcinoma of breast. Histopathology 2006;49:22–34.
22 Banerjee S, Reis-Filho JS, Ashley S, et al. Basal-like breast carcinomas: clinical outcome and response to chemotherapy. J Clin Pathol 2006;59:729–735.
23 Altundag K, Harputluoglu H, Aksoy S, et al. Potential chemotherapy options in the triple negative subtype of breast cancer. J Clin Oncol 2007;25:1294–1295; author reply 1295–1296.
24 Haffty BG, Yang Q, Reiss M, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol 2006;24:5652–5657.
25 Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 2008;26:1275–1281.
26 Rakha EA, Ellis IO, Reis-Filho JS. Immunohistochemical heterogeneity of breast carcinomas negative for estrogen receptors, progesterone receptors and Her2/neu (basal-like breast carcinomas). Mod Pathol 2008;21:1060–1061; author reply 1061–1062.
27 Brenton JD, Carey LA, Ahmed AA, et al. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol 2005;23:7350–7360.
28 Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 2007; 13:2329–2334.
29 Kennecke H, Yerushalmi R, Woods R, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol 2010;28:1684–1691.
30 Thike AA, Cheok PY, Jara-Lazaro AR, et al. Triplenegative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol 2010;23:123–133.
31 Thike AA, Iqbal J, Cheok PY, et al. Triple negative breast cancer: outcome correlation with immunohistochemical detection of basal markers. Am J Surg Pathol 2010;34:956–964.
32 Turner NC, Reis-Filho JS, Russell AM, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 2007;26:2126–2132.
33 Subhawong AP, Subhawong T, Nassar H, et al. Most basal-like breast carcinomas demonstrate the same Rb_/p16+ immunophenotype as the HPV-related poorly differentiated squamous cell carcinomas which they resemble morphologically. Am J Surg Pathol 2009;33:163–175.
34 Jacquemier J, Padovani L, Rabayrol L, et al. Typical medullary breast carcinomas have a basal/myoepithelial phenotype. J Pathol 2005;207:260–268.
35 Vincent-Salomon A, Gruel N, Lucchesi C, et al. Identification of typical medullary breast carcinoma as a genomic sub-group of basal-like carcinomas, a heterogeneous new molecular entity. Breast Cancer Res 2007;9:R24.
36 Bertucci F, Finetti P, Cervera N, et al. Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. Cancer Res 2006;66: 4636–4644.
37 Reis-Filho JS, Milanezi F, Steele D, et al. Metaplastic breast carcinomas are basal-like tumours. Histopathology 2006;49:10–21.
38 Weigelt B, Kreike B, Reis-Filho JS. Metaplastic breast carcinomas are basal-like breast cancers: a genomic profiling analysis. Breast Cancer Res Treat 2009;117:273–280.
39 Lae M, Freneaux P, Sastre-Garau X, et al. Secretory breast carcinomas with ETV6-NTRK3 fusion gene belong to the basal-like carcinoma spectrum. Mod Pathol 2009;22:291–298.
40 Azoulay S, Lae M, Freneaux P, et al. KIT is highly expressed in adenoid cystic carcinoma of the breast, a basal-like carcinoma associated with a favorable outcome. Mod Pathol 2005;18: 1623–1631.
41 Fadare O, Wang SA, Hileeto D. The expression of cytokeratin 5/6 in invasive lobular carcinoma of the breast: evidence of a basal-like subset? Hum Pathol 2008;39:331–336.
42 Fulford LG, Reis-Filho JS, Ryder K, et al. Basal-like grade III invasive ductal carcinoma of the breast: patterns of metastasis and long-term survival. Breast Cancer Res 2007;9:R4.
43 Arnes JB, Brunet JS, Stefansson I, et al. Placental cadherin and the basal epithelial phenotype of BRCA1-related breast cancer. Clin Cancer Res 2005; 11:4003–4011.
44 Pinilla SM, Honrado E, Hardisson D, et al. Caveolin-1 expression is associated with a basal-like phenotype in sporadic and hereditary breast cancer. Breast Cancer Res Treat 2006;99:85–90.
45 Savage K, Leung S, Todd SK, et al. Distribution and significance of caveolin 2 expression in normal breast and invasive breast cancer: an immunofluorescence and immunohistochemical analysis. Breast Cancer Res Treat 2008;110:245–256.
46 Parry S, Savage K, Marchio C, et al. Nestin is expressed in basal-like and triple negative breast cancers. J Clin Pathol 2008;61:1045–1050.
47 Moyano JV, Evans JR, Chen F, et al. AlphaB-crystallin is a novel oncoprotein that predicts poor clinical outcome in breast cancer. J Clin Invest 2006; 116:261–270.
48 Sitterding SM, Wiseman WR, Schiller CL, et al. AlphaB-crystallin: a novel marker of invasive basallike and metaplastic breast carcinomas. Ann Diagn Pathol 2008;12:33–40.
49 Hasegawa M, Hagiwara S, Sato T, et al. CD109, a new marker for myoepithelial cells of mammary, salivary, and lacrimal glands and prostate basal cells. Pathol Int 2007;57:245–250.
50 Hasegawa M, Moritani S, Murakumo Y, et al. CD109 expression in basal-like breast carcinoma. Pathol Int 2008;58:288–294.
51 Reis-Filho J, Pinheiro C, Lambros M, et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol 2006;209: 445–453.
52 Gilbert JA, Goetz MP, Reynolds CA, et al. Molecular analysis of metaplastic breast carcinoma: high EGFR copy number via aneusomy. Mol Cancer Ther 2008; 7:944–951.
53 Rakha E, Ellis I, Reis-Filho J. Are triple-negative and basal-like breast cancer synonymous? Clin Cancer Res 2008;14:618; author reply 618–619.
54 Reis-Filho JS, Savage K, Lambros MB, et al. Cyclin D1 protein overexpression and CCND1 amplification in breast carcinomas: an immunohistochemical and chromogenic in situ hybridisation analysis. Mod Pathol 2006;19:999–1009.
55 Gauthier ML, Berman HK, Miller C, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell 2007; 12:479–491.
56 Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology 2008;52:108–118.
57 Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 2009;15:907–913.
58 Molyneux G, Geyer FC, Magnay FA, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 2010;7:403–417.
59 Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007;13:4429–4434.
60 Rakha EA, El-Sayed ME, Green AR, et al. Prognostic markers in triple-negative breast cancer. Cancer 2007;109:25–32.
61 Tischkowitz M, Brunet JS, Begin LR, et al. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer 2007;7:134.
62 Bauer KR, Brown M, Cress RD, et al. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California Cancer Registry. Cancer 2007;109:1721–1728.
63 Harris LN, Broadwater G, Lin NU, et al. Molecular subtypes of breast cancer in relation to paclitaxel response and outcomes in women with metastatic disease: results from CALGB 9342. Breast Cancer Res 2006;8:R66.
64 Morris GJ, Naidu S, Topham AK, et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: a singleinstitution compilation compared with the National Cancer Institute’s Surveillance, Epidemiology, and End Results database. Cancer 2007;110:876–884.
65 Schneider BP, Winer EP, Foulkes WD, et al. Triplenegative breast cancer: risk factors to potential targets. Clin Cancer Res 2008;14:8010–8018.
66 Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 2007;25:118–145.
67 Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 2010;28: 2784–2795.
68 Rakha EA, Tan DS, Foulkes WD, et al. Are triplenegative tumours and basal-like breast cancer synonymous? Breast Cancer Res 2007;9:404; author reply 405.
69 Kreike B, van Kouwenhove M, Horlings H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res 2007;9:R65.
70 Bertucci F, Finetti P, Cervera N, et al. How basal are triple-negative breast cancers? Int J Cancer 2008; 123:236–240.
71 Calza S, Hall P, Auer G, et al. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res 2006;8:R34.
72 Jumppanen M, Gruvberger-Saal S, Kauraniemi P, et al. Basal-like phenotype is not associated with patient survival in estrogen-receptor-negative breast cancers. Breast Cancer Res 2007;9:R16.
73 Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res 2005; 11:5678–5685.
74 de Ronde JJ, Hannemann J, Halfwerk H, et al. Concordance of clinical and molecular breast cancer subtyping in the context of preoperative chemotherapy response. Breast Cancer Res Treat 2010;119: 119–126.
75 Weigelt B, Mackay A, A’Hern R, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol 2010;11: 339–349.
76 Hu Z. Insight into microRNA regulation by analyzing the characteristics of their targets in humans. BMC Genomics 2009;10:594.
77 Doane AS, Danso M, Lal P, et al. An estrogen receptornegative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 2006;25: 3994–4008.
78 Farmer P, Bonnefoi H, Becette V, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene 2005;24:4660–4671.
79 Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 2009;69:4116–4124.
80 Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 2007;8:R76.
81 Tan DS, Marchio C, Jones RL, et al. Triple negative breast cancer: molecular profiling and prognostic impact in adjuvant anthracycline-treated patients. Breast Cancer Res Treat 2008;111:27–44.
82 Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 2004; 4:814–819.
83 Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene 2006;25: 5846–5853.
84 Foulkes WD, Brunet JS, Stefansson IM, et al. The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloid-microvascular-proliferation+) phenotype of BRCA1-related breast cancer. Cancer Res 2004;64:830–835.
85 Vaziri SA, Krumroy LM, Elson P, et al. Breast tumor immunophenotype of BRCA1-mutation carriers is influenced by age at diagnosis. Clin Cancer Res 2001;7:1937–1945.
86 Foulkes WD, Stefansson IM, Chappuis PO, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 2003; 95:1482–1485.
87 Lakhani SR, Reis-Filho JS, Fulford L, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res 2005;11:5175–5180.
88 Vaziri SA, Tubbs RR, Darlington G, et al. Absence of CCND1 gene amplification in breast tumours of BRCA1 mutation carriers. Mol Pathol 2001;54: 259–263.
89 Palacios J, Honrado E, Osorio A, et al. Phenotypic characterization of BRCA1 and BRCA2 tumors based in a tissue microarray study with 37 immunohistochemical markers. Breast Cancer Res Treat 2005; 90:5–14.
90 Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer 2006;45:1033–1040.
91 Jones C, Ford E, Gillett C, et al. Molecular cytogenetic identification of subgroups of grade III invasive ductal breast carcinomas with different clinical outcomes. Clin Cancer Res 2004;10:5988–5997.
92 van Beers EH, van Welsem T, Wessels LF, et al. Comparative genomic hybridization profiles in human BRCA1 and BRCA2 breast tumors highlight differential sets of genomic aberrations. Cancer Res 2005;65:822–827.
93 Wessels LF, van Welsem T, Hart AA, et al. Molecular classification of breast carcinomas by comparative genomic hybridization: a specific somatic genetic profile for BRCA1 tumors. Cancer Res 2002;62:7110–7117.
94 Melchor L, Honrado E, Garcia MJ, et al. Distinct genomic aberration patterns are found in familial breast cancer associated with different immunohistochemical subtypes. Oncogene 2008;27:3165–3175.
95 Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 2000;92:564–569.
96 Osin P, Lu YJ, Stone J, et al. Distinct genetic and epigenetic changes in medullary breast cancer. Int J Surg Pathol 2003;11:153–158.
97 Beger C, Pierce LN, Kruger M, et al. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA 2001;98:130–135.
98 Welcsh PL, Lee MK, Gonzalez-Hernandez RM, et al. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci USA 2002; 99:7560–7565.
99 Hosey AM, Gorski JJ, Murray MM, et al. Molecular basis for estrogen receptor alpha deficiency in BRCA1-linked breast cancer. J Natl Cancer Inst 2007; 99:1683–1694.
100 Gorski JJ, James CR, Quinn JE, et al. BRCA1 transcriptionally regulates genes associated with the basal-like phenotype in breast cancer. Breast Cancer Res Treat 2010;122:721–731.
101 Bryan BB, Schnitt SJ, Collins LC. Ductal carcinoma in situ with basal-like phenotype: a possible precursor to invasive basal-like breast cancer. Mod Pathol 2006;19:617–621.
102 Paredes J, Lopes N, Milanezi F, et al. P-cadherin and cytokeratin 5: useful adjunct markers to distinguish basal-like ductal carcinomas in situ. Virchows Arch 2007;450:73–80.
103 Tamimi RM, Baer HJ, Marotti J, et al. Comparison of molecular phenotypes of ductal carcinoma in situ and invasive breast cancer. Breast Cancer Res 2008; 10:R67.
104 Vincent-Salomon A, Lucchesi C, Gruel N, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res 2008;14:1956–1965.
105 Dabbs DJ, Chivukula M, Carter G, et al. Basal phenotype of ductal carcinoma in situ: recognition and immunohistologic profile. Mod Pathol 2006; 19:1506–1511.
106 Meijnen P, Peterse JL, Antonini N, et al. Immunohistochemical categorisation of ductal carcinoma in situ of the breast. Br J Cancer 2008;98:137–142.
107 Hannemann J, Velds A, Halfwerk JB, et al. Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Res 2006;8:R61.
108 Shin SJ, Simpson PT, Da Silva L, et al. Molecular evidence for progression of microglandular adenosis (MGA) to invasive carcinoma. Am J Surg Pathol 2009;33:496–504.
109 Khalifeh IM, Albarracin C, Diaz LK, et al. Clinical, histopathologic, and immunohistochemical features of microglandular adenosis and transition into in situ and invasive carcinoma. Am J Surg Pathol 2008;32: 544–552.
110 Salarieh A, Sneige N. Breast carcinoma arising in microglandular adenosis: a review of the literature. Arch Pathol Lab Med 2007;131:1397–1399.
111 Acs G, Simpson JF, Bleiweiss IJ, et al. Microglandular adenosis with transition into adenoid cystic carcinoma of the breast. Am J Surg Pathol 2003;27: 1052–1060.
112 Koenig C, Dadmanesh F, Bratthauer GL, et al. Carcinoma arising in microglandular adenosis: an immunohistochemical analysis of 20 intraepithelial and invasive neoplasms. Int J Surg Pathol 2000;8:303–315.
113 Geyer FC, Kushner YB, Lambros MB, et al. Microglandular adenosis or microglandular adenoma? A molecular genetic analysis of a case associated with atypia and invasive carcinoma. Histopathology 2009;55:732–743.
114 Abd El-Rehim DM, Pinder SE, Paish CE, et al. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol 2004;203:661–671.
115 Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med 2006;355:560–569.
116 Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. Pathology 2009;41:40–47.
117 Tsuda H, Takarabe T, Hasegawa F, et al. Large, central acellular zones indicating myoepithelial tumor differentiation in high-grade invasive ductal carcinomas as markers of predisposition to lung and brain metastases. Am J Surg Pathol 2000;24:197–202.
118 Hicks DG, Short SM, Prescott NL, et al. Breast cancers with brain metastases are more likely to be estrogen receptor negative, express the basal cytokeratin CK5/ 6, and overexpress HER2 or EGFR. Am J Surg Pathol 2006;30:1097–1104.
119 Rodriguez-Pinilla SM, Sarrio D, Honrado E, et al. Prognostic significance of basal-like phenotype and fascin expression in node-negative invasive breast carcinomas. Clin Cancer Res 2006;12:1533–1539.
120 Smid M, Wang Y, Zhang Y, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res 2008;68:3108–3114.
121 Marchio C, Weigelt B, Reis-Filho JS. Adenoid cystic carcinomas of the breast and salivary glands (or ‘the strange case of Dr Jekyll and Mr Hyde’ of exocrine gland carcinomas). J Clin Pathol 2010;63:220–228.
122 Keam B, Im SA, Kim HJ, et al. Prognostic impact of clinicopathologic parameters in stage II/III breast cancer treated with neoadjuvant docetaxel and doxorubicin chemotherapy: paradoxical features of the triple negative breast cancer. BMC Cancer 2007; 7:203.
123 Sanchez-Munoz A, Garcia-Tapiador AM, Martinez- Ortega E, et al. Tumour molecular subtyping according to hormone receptors and HER2 status defines different pathological complete response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. Clin Transl Oncol 2008;10: 646–653.
124 Sirohi B, Arnedos M, Popat S, et al. Platinum-based chemotherapy in triple-negative breast cancer. Ann Oncol 2008;19:1847–1852.
125 Teschendorff AE, Miremadi A, Pinder SE, et al. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol 2007;8:R157.
126 Reyal F, van Vliet MH, Armstrong NJ, et al. A comprehensive analysis of prognostic signatures reveals the high predictive capacity of the proliferation, immune response and RNA splicing modules in breast cancer. Breast Cancer Res 2008;10:R93.
127 Teschendorff AE, Caldas C. A robust classifier of high predictive value to identify good prognosis patients in ER-negative breast cancer. Breast Cancer Res 2008; 10:R73.
128 Desmedt C, Haibe-Kains B, Wirapati P, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res 2008;14:5158–5165.
129 Grigoriadis A, Caballero OL, Hoek KS, et al. CT-X antigen expression in human breast cancer. Proc Natl Acad Sci USA 2009;106:13493–13498.
130 Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol 2008;26:3785–3790.
131 Turner N, Tutt A, Ashworth A. Targeting the DNA repair defect of BRCA tumours. Curr Opin Pharmacol 2005;5:388–393.
132 Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009;361:123–134.
133 Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol 2010;28: 2512–2519.
134 Tutt A, Robson M, Garber JE, et al. Oral poly(ADPribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010; 376:235–244.
135 O’Shaughnessy J, Yoffe M, Osborne C, et al. Triple negative breast cancer: a phase 2, multi-center, openlabel, randomized trial of gemcitabine/carboplatin (G/C), with or without BSI-201, a PARP inhibitor. Cancer Res 2009;69:193S-S.
136 Foulkes WD, Reis-Filho JS, Narod SA. Tumor size and survival in breast cancer—a reappraisal. Nat Rev Clin Oncol 2010;7:348–353.
137 Badve S. Does tumor size trump biology? Clin Breast Cancer 2010;10:111–112.
138 Bhargava R, Beriwal S, McManus K, et al. CK5 is more sensitive than CK5/6 in identifying the ‘basallike’ phenotype of breast carcinoma. Am J Clin Pathol 2008;130:724–730.
139 Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63.
140 Ledford H. The death of microarrays? Nature 2008;455:847.
141 Shendure J. The beginning of the end for microarrays? Nat Methods 2008;5:585–587.

华夏病理/粉蓝医疗
为基层医院病理科提供全面解决方案,
努力让人人享有便捷准确可靠的病理诊断服务。
-
本帖最后由 于 2011-02-19 18:35:00 编辑
Figure 1 The typical morphological features of triple-negative/basal-like cancer are those of a high-grade ductal carcinoma (
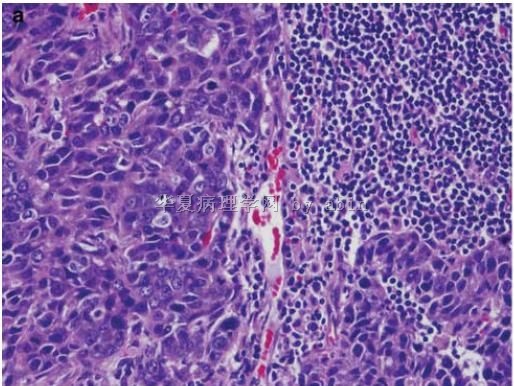
名称:图1
描述:图1
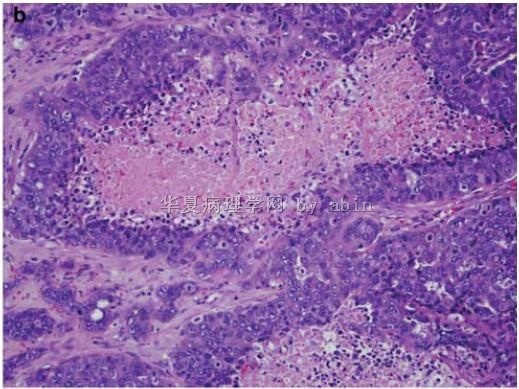
名称:图2
描述:图2
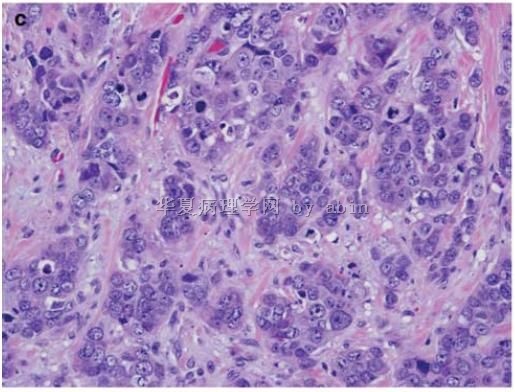
名称:图3
描述:图3
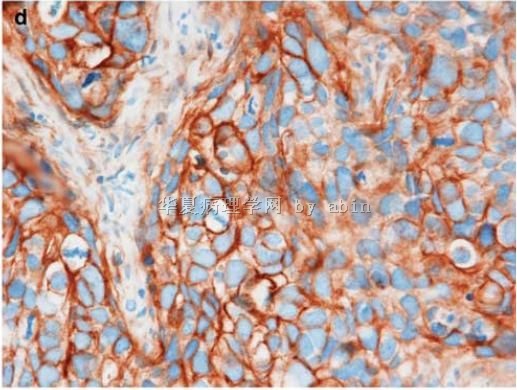
名称:图4
描述:图4
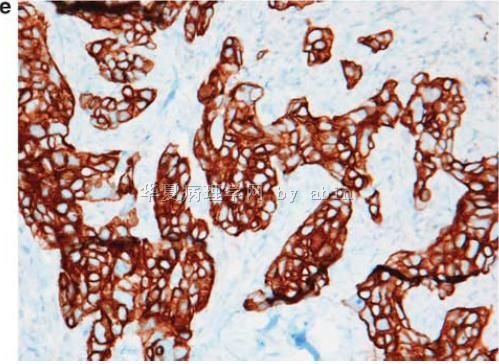
名称:图5
描述:图5

华夏病理/粉蓝医疗
为基层医院病理科提供全面解决方案,
努力让人人享有便捷准确可靠的病理诊断服务。